Phylogenetics¶
Phylogenetics is a minimal Python API for doing phylogenetics. It manages the annoying aspects of phylogenetics (i.e. file conversion) for you and lets you focus on exploring and interpreting the data.
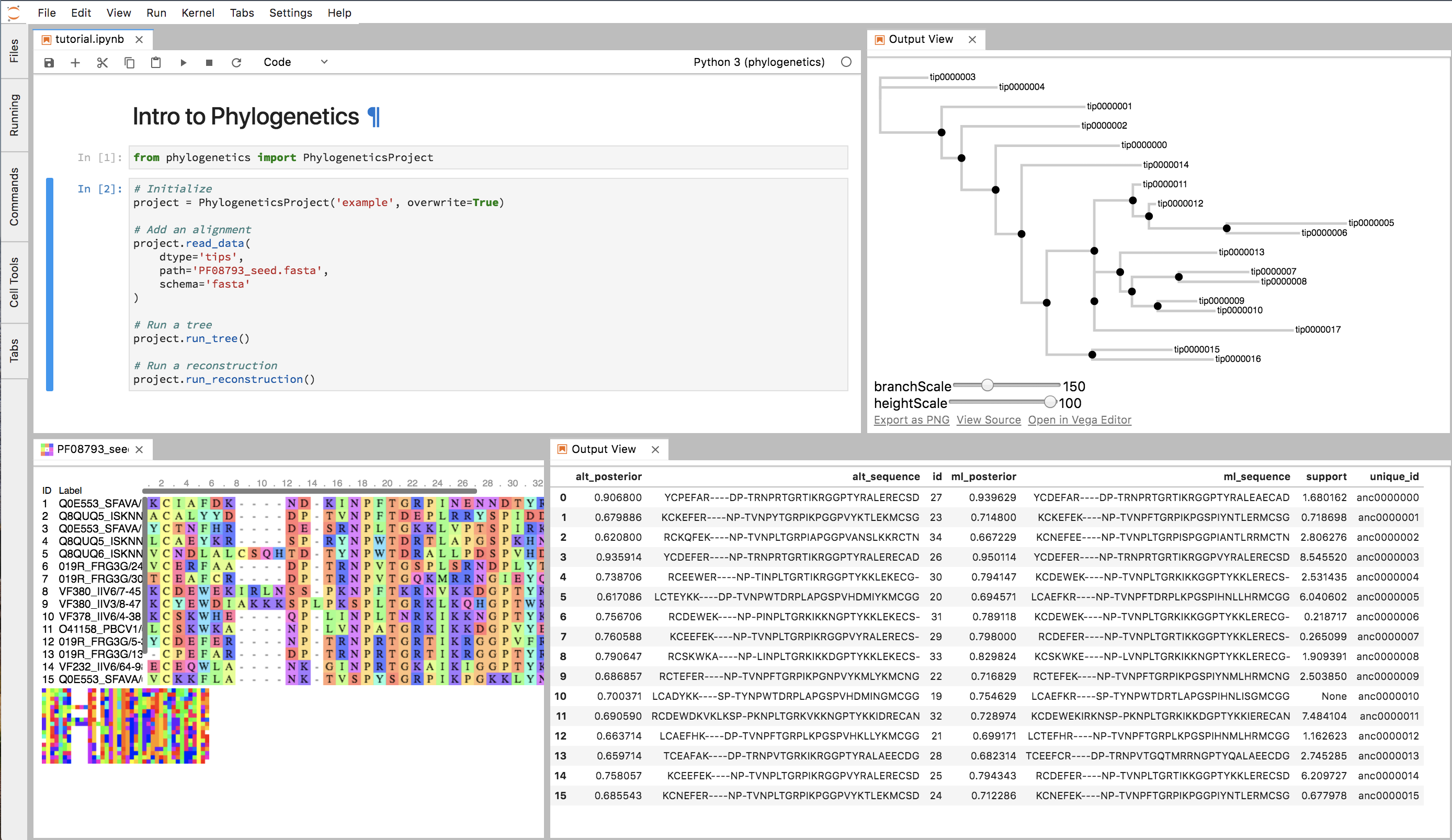
Basic Example¶
The main object in phylogenetics is the PhylogeneticsProject. Start by initializing the PhylogeneticsProject
object, pointing to a directory where you’d like to store all the phylogenetic data/output.
# Imports
from phylogenetics import PhylogeneticsProject
# Initialize a project class
project = PhylogeneticsProject(project_dir='project')
Then, add an alignment (using any file schema you’d like). These sequences will be the tips of your tree. You can now begin building trees and reconstructing ancestral sequences.
project.read_data(dtype='tips', path='alignment.fasta', schema='fasta')
# Run PhyML to construct a phylogenetic
# tree by maximum likelihood.
project.run_tree()
# Reconstruct ancestral sequences using default settings.
project.run_reconstruction()
Installation¶
Install from PyPi:
pip install phylogenetics
To install a development version:
git clone https://github.com/Zsailer/phylogenetics
cd phylogenetics
pip install -e .
Dependencies¶
phylogenetics manages phylogenetics data. Currently, it doesn’t do any of the phylogenetic calculations itself. For this, use external tools like:
phylogenetics is built on top of following python stack:
- DendroPy: A Python library for phylogenetic scripting, simulation, data processing and manipulation.
- ToyTree: A minimalist tree plotting library using toyplot graphs
- PhyloPandas: Pandas for phylogenetics
- PyASR: Ancestral Sequence Reconstruction in Python